Using Tabular Explainers
Introduction
In this tutorial we explain the tabular data classification using 4 different explainers, including LIMETabular,
SHAPKernel, SHAPGradient, and PseudoLinearCoef.
The complete code of the tutorial below is using_tabular_explainers.py.
Import Dataset
We use the Iris dataset for the demonstration. These data sets consist of 3 different types of irises’ petal and sepal lengths.
import sklearn.datasets
import mindspore as ms
iris = sklearn.datasets.load_iris()
# feature_names: ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
feature_names = iris.feature_names
# class_names: ['setosa', 'versicolor', 'virginica']
class_names = list(iris.target_names)
# convert data and labels from numpy array to mindspore tensor
# use the first 100 samples
data = ms.Tensor(iris.data, ms.float32)[:100]
labels = ms.Tensor(iris.target, ms.int32)[:100]
# explain the first sample
inputs = data[:1]
# explain the label 'setosa'(class index 0)
targets = 0
Import Model
Here we define a simple linear classifier.
import numpy as np
import mindspore.nn as nn
class LinearNet(nn.Cell):
def __init__(self):
super(LinearNet, self).__init__()
# input features: 4
# output classes: 3
self.linear = nn.Dense(4, 3, activation=nn.Softmax())
def construct(self, x):
x = self.linear(x)
return x
net = LinearNet()
# load pre-trained parameters
weight = np.array([[0.648, 1.440, -2.05, -0.977], [0.507, -0.276, -0.028, -0.626], [-1.125, -1.183, 2.099, 1.605]])
bias = np.array([0.308, 0.343, -0.652])
net.linear.weight.set_data(ms.Tensor(weight, ms.float32))
net.linear.bias.set_data(ms.Tensor(bias, ms.float32))
Using LIMETabular
LIMETabular approximates the machine learning model with a local, interpretable model to explain each individual
prediction.
from mindspore_xai.explainer import LIMETabular
# convert features to feature stats
feature_stats = LIMETabular.to_feat_stats(data, feature_names=feature_names)
# initialize the explainer
lime = LIMETabular(net, feature_stats, feature_names=feature_names, class_names=class_names)
# explain
lime_outputs = lime(inputs, targets, show=True)
print("LIMETabular:")
for i, exps in enumerate(lime_outputs):
for exp in exps:
print("Explanation for sample {} class {}:".format(i, class_names[targets]))
print(exp, '\n')
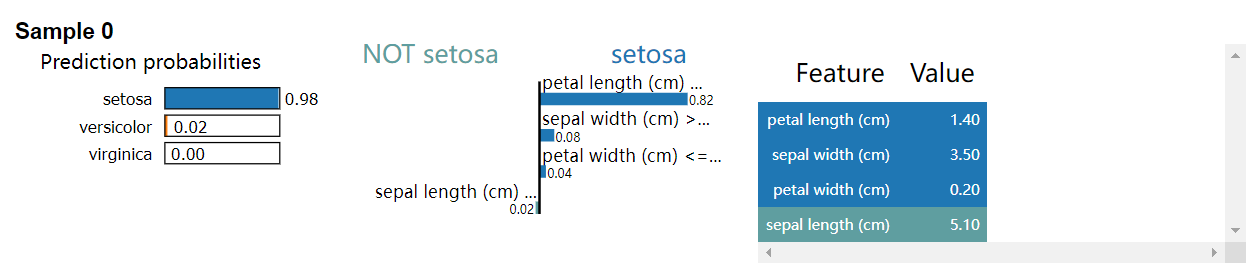
output:
LIMETabular:
Explanation for sample 0 class setosa:
[('petal length (cm) <= 1.60', 0.8182714590301656),
('sepal width (cm) > 3.30', 0.0816516722404966), ('petal width (cm) <= 0.30', 0.03557190104069489),
('sepal length (cm) <= 5.10', -0.021441399016492325)]
LIMETabular also supports a callable function, for example:
def predict_fn(x):
return net(x)
# initialize the explainer
lime = LIMETabular(predict_fn, feature_stats, feature_names=feature_names, class_names=class_names)
Using SHAPKernel
SHAPKernel is a method that uses a special weighted linear regression to compute the importance of each feature.
from mindspore_xai.explainer import SHAPKernel
# initialize the explainer
shap_kernel = SHAPKernel(net, data, feature_names=feature_names, class_names=class_names)
# explain
shap_kernel_outputs = shap_kernel(inputs, targets, show=True)
print("SHAPKernel:")
for i, exps in enumerate(shap_kernel_outputs):
for exp in exps:
print("Explanation for sample {} class {}:".format(i, class_names[targets]))
print(exp, '\n')
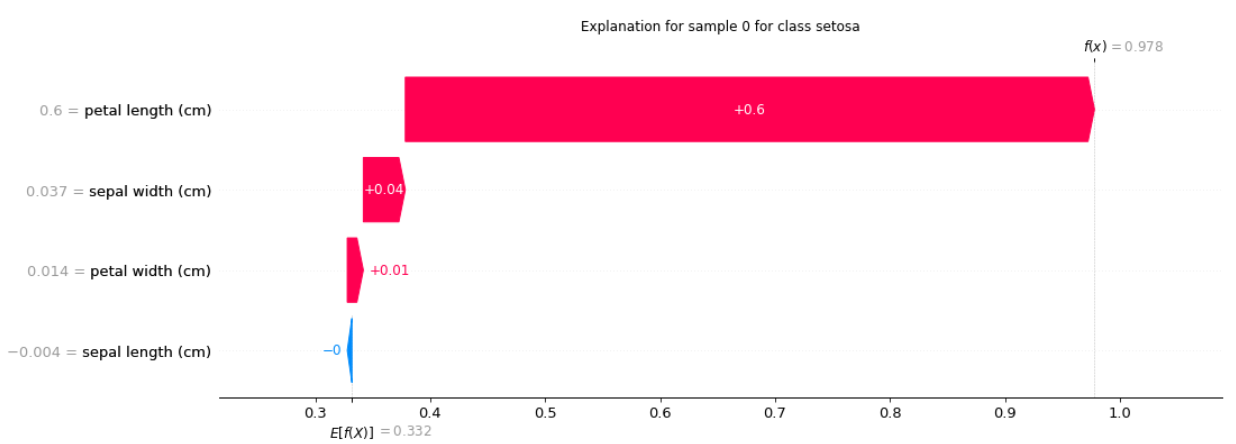
output:
SHAPKernel:
Explanation for sample 0 class setosa:
[-0.00403276 0.03651359 0.59952676 0.01399141]
SHAPKernel also supports a callable function, for example:
# initialize the explainer
shap_kernel = SHAPKernel(predict_fn, data, feature_names=feature_names, class_names=class_names)
Using SHAPGradient
SHAPGradient explains a model using expected gradients (an extension of integrated gradients).
from mindspore_xai.explainer import SHAPGradient
# initialize the explainer
shap_gradient = SHAPGradient(net, data, feature_names=feature_names, class_names=class_names)
# explain
shap_gradient_outputs = shap_gradient(inputs, targets, show=True)
print("SHAPGradient:")
for i, exps in enumerate(shap_gradient_outputs):
for exp in exps:
print("Explanation for sample {} class {}:".format(i, class_names[targets]))
print(exp, '\n')
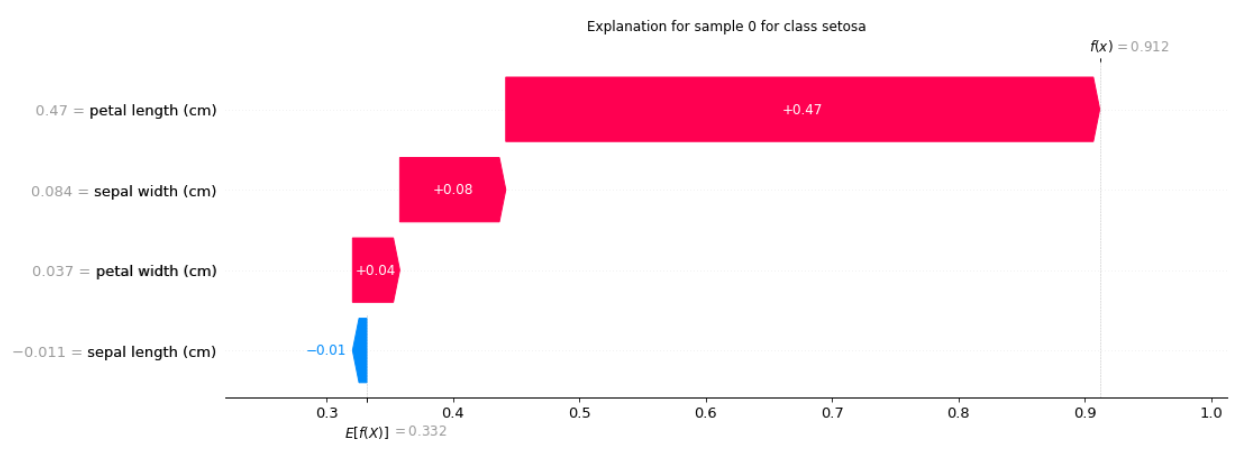
output:
SHAPGradient:
Explanation for sample 0 class setosa:
[-0.0112452 0.08389313 0.47006473 0.0373782]
Using PseudoLinearCoef
PseudoLinearCoef provides a global attribution method to measure the sensitivity of features around the classifier decision boundary.
from mindspore_xai.explainer import PseudoLinearCoef
# initialize the explainer
plc_explainer = PseudoLinearCoef(net, len(class_names), feature_names=feature_names, class_names=class_names)
# explain
plc, relative_plc = plc_explainer(data, show=True)
print("Pseudo Linear Coef.:")
for target, target_name in enumerate(class_names):
print(f"class {target_name}")
print(str(plc[target]))
print("\nRelative Pseudo Linear Coef.:")
for target, target_name in enumerate(class_names):
for view_point, view_point_name in enumerate(class_names):
if target == view_point:
continue
print(f"{target_name} relative to {view_point_name}")
print(str(relative_plc[target, view_point]))
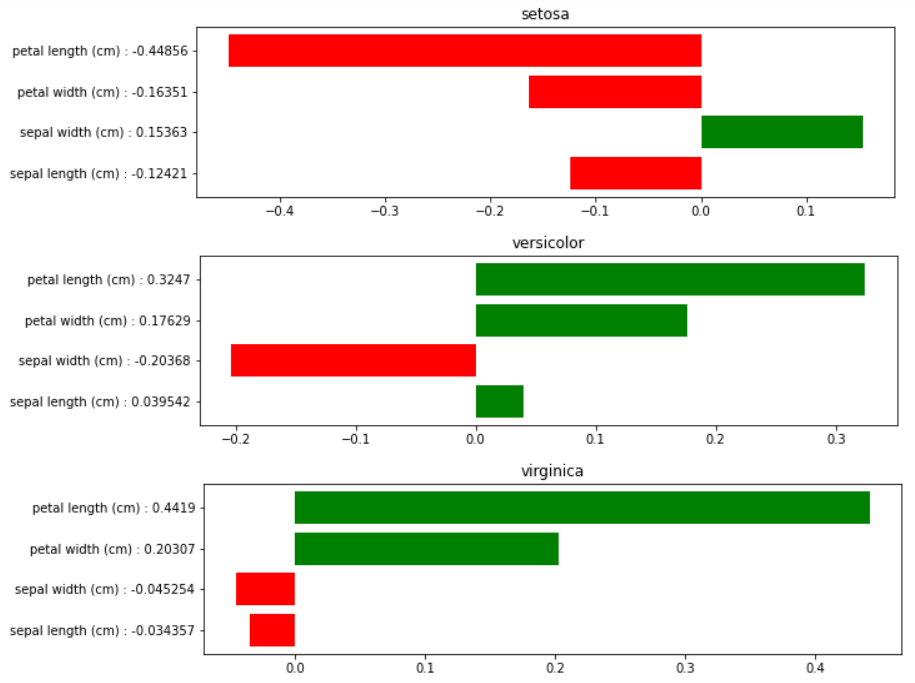
output:
Pseudo Linear Coef.:
class setosa
[-0.12420721 0.15363358 -0.44856226 -0.16351467]
class versicolor
[ 0.03954152 -0.20367564 0.3246966 -0.17629193]
class virginica
[-0.03425665 -0.04525428 0.44189668 0.20307252]
Relative Pseudo Linear Coef.:
setosa relative to versicolor
[-0.12564947 0.15629557 -0.44782427 -0.16126522]
setosa relative to virginica
[-0.11122696 0.12967573 -0.45520434 -0.18375972]
versicolor relative to setosa
[ 0.02240782 -0.23672473 0.3889126 0.21666989]
versicolor relative to virginica
[ 0.21087858 0.1268154 -0.31746316 -0.22748768]
virginica relative to setosa
[ 0.07109872 -0.08392082 0.5585888 0.23082316]
virginica relative to versicolor
[-0.15152863 -0.00229146 0.31223866 0.17223847]